The Google Summer of Code program is ending on August 31, and as a final step I am required to write a report on all the work that I have done in the last three months. That's what I will do in this final blog post, while sharing some behind-the-scenes and some details that I may have missed in the previous posts.
The proposal
Gianluca Della Vedova contacted me back in February to discuss an interesting project idea from the OBF (Open Bioinformatics Foundation) called "The fastest GPU Variation Graph explorer (VG)". I was immediately drawn to the project, even though my parallel programming skills were admittedly limited. One of the project requirements was to use Rust, a programming language I had heard a lot about, but never actually used.
So, after some help from Gianluca, I wrote a proposal, and sent it to my future mentors: Pjotr Prins, Erik Garrison and George Githinji. To be honest, I really did not think my proposal would get accept, but thankfully it was! A few weeks later, I met with Pjotr and the others in an online conference, and we started discussing what I would actually do during GSoC.
Since I had little to no experience in Rust, we decided that re-implementing an already existing tool would be best, so that I would have some kind of reference. The tool we chose was
Flavia95's GFAtoVCF, which while being relatively straightforward, would prove to be a great introduction to the world of Variation Graphs. And so, on June 1, I started working on my Rust re-implementation of GFAtoVCF, called
rs-gfatovcf.
rs-gfatovcf (June - July)
My first contact with Rust was rather... rough. While it looks similar to C++, it feels like a completely different language, mostly due to the
borrow checker. In order to start building some Rust knowledge, I started by mindlessly copying Flavia's script line-by-line, while also trying to make it more Rust-y in the process. While doing this, I noticed that there were multiple references to a library called
Handlegraph, which I had never heard of before. At first I thought I had to re-implement that as well, but thankfully
chfi had already done that with his
rs-handlegraph. This library actually allows for storing Variation Graphs inside memory, and it is essential in a program that essentialy converts a GFA (that is, a Variation Graph) into a VCF.
Once I finished "translating" the code to Rust, I started working on how to actually improve it. First, I wrapped more parts of code into functions, so that they were easier to read; these functions were also splitted across multiple files, to further increase readability. Second, I replaced the DFS with a BFS, since I was getting inconsistent results when using a DFS. Third, I added a lot of tests, to make sure that everything worked correctly.
In the first two weeks of July, after some research on Rust concurrency, I started working on a
concurrent version of rs-gfatovcf, mostly focused around the
Rayon library. This however proved to be rather difficult, mostly due to how complex rs-gfatovcf is (and to me being relatively inexperienced with Rust and parallelism). I plan on returning to work on parallelism once I have a greater understanding on how it works in Rust.
Even with the sequential version of rs-gfatovcf up and running, I noticed that in some instances, nested bubbles (also known as
superbubbles) were not being correctly detected. Addressing this issue required a complete overhaul of the bubble detection process, and Erik suggested taking a look at a paper called
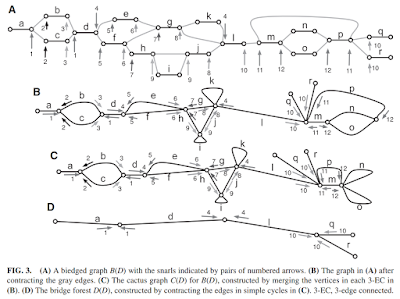
Superbubbles, Ultrabubbles and Cacti by Benedict Paten et al. which contained a more refined approach for bubble detection.
rs-cactusgraph (August)
In the final section of that paper, the author stated that the approach was already implemented within vg, so I decided to take a look at
vg's source code. I quickly found the related files, and initially decided to re-implement that in Rust as well. It didn't take long before I noticed that the implementation relied on another library called
sonLib (or rather,
pinchesAndCacti) which would have required too much time to re-implement. So I dropped the idea of a "translation" and instead decided to create my own implementation of the Superbubble detection algorithm, in a repository called
rs-cactusgraph.
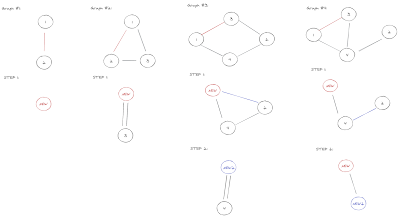
The first decision I had to make was which data structure to use. Handlegraph was the obvious choice, but I couldn't really get it work with the algorithm, so I decided to make my own implementation, backed by
Petgraph. I therefore created a BiedgedGraph class and a way to convert a Handlegraph into a BiedgedGraph, so that I can run the algorithm on any graph from rs-gfatovcf.
When implementing the algorithm I tried to stay as close as possible to the original description in the paper. The hardest step to implement was the second one, that is merging all nodes in each
3-connected-component of the graph. Thankfully, chfi came to the rescue (again) with
rs-3-edge, so all I had to do was to create an interface between his code and mine.
As of now, the whole algorithm has been implemented, but some more testing is definitely required. I also have to find a way to map the Cactus Graph back to the Handlegraph, in order to obtain the bubbles, which will be used in the variant identification phase of rs-gfatovcf.
Future work
The first thing that needs to be done is finishing up rs-cactusgraph, and making sure it works correctly. Once that is done, it needs to be integrated inside rs-gfatovcf which shouldn't take too long, and the resulting VCF should be validated against known tools such as vg deconstruct. This should obviously be repeated for as many graphs as possible, just to be as sure as possible that it works correctly.
Another interesting future project would be to complete the concurrent version, and actually start looking into parallelism. I really need to read more on these topics (both in general and with respect to Rust specifically), but once I do that I will definitely come back to this project.
Why this project matters
Due to its current international relavance, we are especially interested in finding variants (that is, a VCF file) from
COVID-19 pangenomes, as part of the
COVID-19 PubSeq Project. Maybe by looking at variants we will be able to understand how many different strains of the virus are currently circulating, or maybe we will be able to create ad-hoc treatments for each patient, according to how similar or different the virus samples extracted from him/her are to other known genomes.
TL;DR
- rs-gfatovcf is a tool that finds variants in a Variation Graph. I worked on it in June and July, and with respect to the original implementation it is 100% done. You can find it at: https://github.com/HopedWall/rs-gfatovcf
- rs-cactusgraph represents an attempt at improving rs-gfatovcf's bubble detection capabilities. I worked on it in August, and it's about 70% done. You can find it at: https://github.com/HopedWall/rs-cactusgraph
- This tool will be used to analyze COVID-19 pangenomes, and by leveraging parallel computing we will be able to do so much faster.
Acknowledgments
I want to thank:
Also a big thank you to OBF and Google for allowing me to take part in this program!